Current Projects & Research Themes
The Nutritional Ecology of Human Health & Ageing
The Nutritional Ecology of Human Health & Ageing
The Nutritional Ecology of Human Health & Ageing

How does what we eat affect our lifespan and healthspan? This is the question everyone wants to answer. Unfortunately this is one of the hardest questions in all of nutrition science. To try and answer these questions we take an ecological approach to analysing nutritional epidemiology datasets.
For example, we have been using the 'Geometric-Framework for Nutrition' (see below) to assess to national nutrient supplies associate with the risk of mortality across the human life-course.
Gene-Diet Interactions in Fruit-Fly
The Nutritional Ecology of Human Health & Ageing
The Nutritional Ecology of Human Health & Ageing
The fruit-fly, Drosophila melanogaster, is arguably the most widely used model in the study of genetics. We use fruit-fly research to identify novel gene-diet interactions and diet-responsive pathways that govern development, ageing and reproductive function.
Our research has shown, for example, that the JNK-pathway (a stress-signalling pathway) affects ability to tolerate high-sugar diets through its effects on de-novo lipogenesis.
The Evolution of Ageing
The Nutritional Ecology of Human Health & Ageing

An organism that reproduces in perpetuity will be infinitely more fit than one that dies. Why then has evolution left so many organisms with a life-history trait that increases the risk of death with time ... ageing.
Is ageing inevitable? How have some organisms seemingly evolved their way out of ageing? Could ageing even be adaptive somehow? Could the stability of the nutritional environment play a role? These are the types of questions we try and answer using theoretical modelling and literature synthesis.
The Geometric Framework for Nutrition
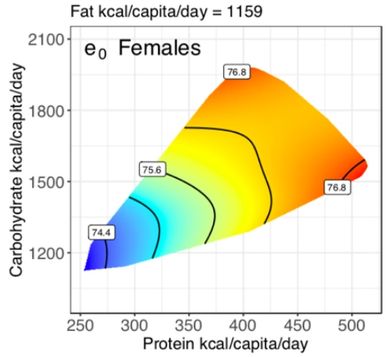
The Geometric Framework for Nutrition (GFN) is a conceptual and analytical framework for separating the interactive effects of different nutrients on health, appetite and life-history. It can be used to design experiments and statistical analyses.
Originally developed by collaborators Stephen Simpson and David Raubenheimer, the GFN is now used widely in evolutionary ecology and increasingly in human nutrition. Our group has lead the way in developing analytical workflows for the GFN, as well as in applying the GFN to problems in organismal life history.
The Protein-Leverage Hypothesis

Emerging data suggest that the quantity of protein eaten is a powerful determinant of satiety. As a consequence, we may have to consume a lot of a food with low-protein content before we feel like we have eaten enough protein to feel full. If this is true, modern nutritional environments that are rich in energy-dense and protein-dilute foods may drive in overconsumption of energy and promote obesity. This is the protein-leverage hypothesis.
We have been developing models and workflows that allow the user to test the protein-leverage hypothesis. Working with our collaborators we have also been using these methods to test the hypothesis with epidemiological data.
Meta-Analysis
The amount of data in the primary literature is growing more and more every day. How are we supposed to stay on top of all of these studies, let alone meaningfully compare their findings?
Meta-analysis is a statistical tool that can be used to combine and contrast the results of all the individual studies on a given topic.
We have a great deal of expertise with this approach, including the development of novel methods. A major innovation was our development of methods for meta-analysis of variability. With these tools the user can assess how a treatment (e.g., diet) affects variation in a traits of interest.
Research Partners & Affiliations
Charles Perkins Centre

The Charles Perkins Centre (CPC) is a multidisciplinary research centre at the University of Sydney. The CPC is dedicated to easing the burden of chronic and non-communicable disease. The CPC Hub on the Camperdown campus is the home of our research.
Learn more here: https://www.sydney.edu.au/charles-perkins-centre/
GECKO Consortium

We work closely with, and have members who are part of, the GECKO, or Gametic Epigenetics Consortium against Obesity, consortium. GECKO is a consortium of researchers interested in nutrition, epigenetic inheritance and metabolic health.
Learn more here: https://geckoconsortium.com/
Sydney Precision Data Science Centre
Sydney Precision Data Science Centre

The Sydney Precision Data Science Centre (SPDS) was founded at the University of Sydney in 2022, and we are among its founding members. SPDS provides a research hub to all interdisciplinary researchers in data-intensive science.
Learn more here: https://www.sydney.edu.au/science/our-research/research-centres/sydney-precision-data-science-centre.html
Precision Nutrition Research Group
Sydney Precision Data Science Centre

Led by Professor Stephen Simpson, the Precision Nutrition Research Group emphasises 'bottom-up' research, and is especially concerned with the way in which lower-level processes lead to higher level outcomes. We work closely with them in the analysis of pre-clinical mouse data, epidemiological data and in many other domains.
Learn more here: https://www.sydney.edu.au/science/about/our-people/academic-staff/stephen-simpson.html
Islet Biology and Metabolism Research Group

Led by Dr Melkam Kebede, this group is exploring how pancreatic beta-cells work in the management of insulin. Insulin is one of the mechanistic regulators of metabolism, and ultimately life-history. We have overlapping group members, and working together on mapping the insulin granule proteome.
Learn more here: https://www.sydney.edu.au/charles-perkins-centre/our-research/research-groups/islet-biology-metabolism-research-group.html
I-DEEL

The Inter-Disciplinary Ecology and Evolution Lab (I-DEEL) is at the University of New South Wales. We have been working most closely with them in the development of new methods for meta-analysis. The group is led by Professor Shinchi Nakagawa.
Learn more here: http://www.i-deel.org/
Nutritional Ecology Research Group
Nutritional Ecology Research Group
Nutritional Ecology Research Group

We work closely with the Nutritional Ecology Research Group led by Professor David Raubenheimer. This group has several overlapping research interests with our own. We have been working closely with them to establish analytical solutions to complex problems in nutrition.
Learn more here: https://www.sydney.edu.au/science/about/our-people/academic-staff/david-raubenheimer.html
Metabolic Cybernetics Group
Nutritional Ecology Research Group
Nutritional Ecology Research Group

The metabolic Cybernetics group devises quantitative strategies to understand how cells or organisms respond acutely to stresses such as meals or exercise and how such changes are modulated by environmental versus genetic diversity. We have been working with them on projects ranging from cell signalling to gene-diet interactions in mice. The group is led by Professor David James.
Learn more here: https://metabocybernetics.com/
Useful Links & Resources
Al's Github Page
Our Instagram Page
Our Instagram Page
Find (hopefully) useful R code and data associated with our research here.
Our Instagram Page
Our Instagram Page
Our Instagram Page
A link to our social.
This website uses cookies.
We use cookies to analyze website traffic and optimize your website experience. By accepting our use of cookies, your data will be aggregated with all other user data.